Relevant Specification Extracts:
OCR
6.2.2 Amino acids
Learning outcomes
Learners should be able to demonstrate and apply their knowledge and understanding of:
Reactions of amino acids
The general formula for an α-amino acid as RCH(NH2)COOH and the following reactions of amino acids:
(i) reaction of the carboxylic acid group with alkalis and in the formation of esters
(ii) reaction of the amine group with acids
Edexcel 18B
16. understand the properties of 2-amino acids, including:
i acidity and basicity in solution, as a result of the formation of zwitterions
ii effect of aqueous solutions on plane-polarised monochromatic light
17. understand that the peptide bond in proteins:
i is formed when amino acids combine, by condensation polymerisation
ii can be hydrolysed to form the constituent amino acids, which can be separated by chromatography
AQA
3.3.13 Amino acids and proteins
Amino acids and proteins are the molecules of life. In this section, the structure and bonding in these molecules and the way they interact is studied. Drug action is also considered.
3.3.13.1 Amino acids (A-level only)
Amino acids have both acidic and basic properties, including the formation of zwitterions.
Students should be able to draw the structures of amino acids as zwitterions and the ions formed from amino acids:
in acid solution and in alkaline solution.
3.3.13.2 Proteins (A-level only)
Proteins are sequences of amino acids joined by peptide links.
The importance of hydrogen bonding and sulfur–sulfur bonds in proteins.
The primary, secondary (α-helix and β–pleated sheets) and tertiary structure of proteins.
Hydrolysis of the peptide link produces the constituent amino acids.
Amino acids can be separated and identified by thin-layer chromatography.
Amino acids can be located on a chromatogram using developing agents such as ninhydrin or ultraviolet light and identified by their Rf values.
Students should be able to:
- draw the structure of a peptide formed from up to three amino acids
- draw the structure of the amino acids formed by hydrolysis of a peptide
- identify primary, secondary and tertiary structures in diagrams
- explain how these structures are maintained by hydrogen bonding and S–S bonds
- calculate Rf values from a chromatogram.
Amino acids and Proteins
In this third post on condensation polymers, I’m looking at the properties of amino acids and the formation of proteins from amino acids, defining the repeat units of proteins and explaining the difference between primary, secondary, tertiary and quaternary structures of proteins, and the nature of the intermolecular forces that exist between these polymer molecules.
I’m going to discuss how amino acids combine with each other to form protein chains and how to spot the repeat unit and the peptide linkage between repeat units of protein.
Amino Acids
As their name suggests amino acids are part of both the amine family of molecules and the carboxylic acid family contain the –COOH and the –NH2 functional groups.
They have the general formula RCH2(NH2)COOH
You will also notice that amino acids are called α–aminoacids or 2–aminocarboxylic acids because the –NH2 group is on the second carbon atom of the chain.
Amino acids as optically active
You will also see that there is the potential for amino acid molecules to contain four different functional groups on the second carbon atom.
Four different functional groups on the second carbon atom means that the amino acid is optically active. It has two enantiomers: they rotate the plan of plane polarised light in opposite directions.
You can see the effect here in these diagrams:
Amino acids as zwitterions
Amino acids then carry the properties of both the carboxylic acid group–they are weak acids and the amino group–they are weak bases. In fact they exist in an ionic form called a zwitterion (from the German for twin: zwitter). In this form the proton from the carboxylic acid group has migrated to the amino group. You can see this in the equation below:
Reaction with alcohols to form esters
Amino acids react with alkalis to form salts and with acids to form salts
Polymerisation of amino acids
Amino acids are the monomers of protein. Amino acids combine in a condensation reaction i.e. the addition of two amino acids followed by the elimnation of a molecule of water.
The resultant linkage between the two amino acid residues is called a peptide link.
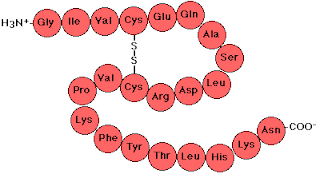
The basic protein structure is called its Primary Structure and this is just a sequence of amino acid residues. See below and note the peptide linkages:
Note that is writing out the primary structure we begin with the amino nitrogen on the left.
However, amino acid R groups can interact with one another under the appropriate circumstances and this results in two types of secondary structure called α–helix and β–pleated sheet. These two secondary structures are illustrated below.
You should be able to recognise and identify these structures in diagrams.
The structures are held in place when hydrogen bonds form between amino acid side chains or disulphide bridges form between cysteine side chains. These are illustrated below:
When the secondary structures interact the hydrophobic water–hating side chains tend to move to the centre of the protein molecule and the water–soluble hydrophilic side chains move to the surface of the protein. This produces a third structure type of structure obviously called the protein tertiary structure.
The α–helix and β–pleated sheet are like the struts and plates that hold the tertiary structure in place.
You can see the tertiary structure of myoglobin below.
Hydrolysis of a protein and analysis of its hydrolysate
To analyse a protein and determine the composition of a protein’s primary structure, the first step is to hydrolyse the molecule.
Hydrolysis is usually accomplished refluxing the protein for several hours in 3M HCl i.e. acid hydrolysis. The resultant hydrolysate is then neutralised.
Hydrolysis occurs at the peptide linkage.see below
Paper or thin layer chromatography can be used to determine which amino acids are present in the hydrolysate. See the diagram below:
Comparing known amino acid Rf values with those of the components of the hydrolysate will provide the evidence needed to identify the amino acids in the protein’s primary structure. See the illustration below:
How to determine an Rf value.
Below are results of using ninhydrin to spot the movement of the amino acids on the chromatogram
Summary of Protein Structure